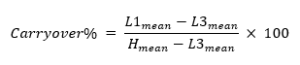The latest study added to Validation Manager is the Sample Carryover study. The purpose of the study is to find out whether a specimen is somehow contaminated by the previous one, leading to low-level samples run directly after high-level samples to give too high results.
The challenge in building the study was that there’s a wide variety of protocols used for this purpose. We wanted to build a study that would cater to the needs of most of our users, but no single protocol can do this. As a result, we now have a study with two optional modes, which should cover most protocols used for this purpose. If you cannot conduct your sample carryover protocol with this study, drop us a message describing your protocol and a reference on which it’s based, and we’ll see how to make it work.
Mode 1: The Cofrac protocol
Cofrac SH GTA 04 suggests a protocol where three high-level samples (H1, H2, H3) are run before three low-level samples (L1, L2, L3), and this sequence is repeated five times. Relative carryover is then calculated as
where L1mean is the mean of the results of all the low-level samples run immediately after the high-level samples, L3mean is the mean of the results of all the low-level samples run last in a sequence, and Hmean is the mean of the results of all the high-level samples run in this protocol.
To make this mode suit also to a bit different protocols, we made this protocol customizable in two ways.
First, you can freely plan your protocol, how many high samples and how many low samples each sequence should contain, and how many times the sequence should be repeated. If you are running your verification using the Cofrac protocol, you would plan to have three high-level samples and three low-level samples run five times. But you can just as well have for example a sequence of two high-level samples and four low-level samples, each sequence run ten times.
Second, the calculations are based on sample categories. After importing your results, you should drag all your samples to the correct categories:
-
High Level Samples: samples in this category are used in calculating Hmean
-
Low Test Samples: samples in this category are used in calculating L1mean
-
Low Control Samples: samples in this category are used in calculating L3mean
-
Other: samples in this category are not used in calculations, but you can see their results on the report to visually assess if their results are on the expected level, and whether the protocol you measured matches the sequence you were supposed to measure.
When following the Cofrac protocol, you would categorize all your high-level samples as High Level Samples. All the low-level samples run directly after high-level samples would be categorized as Low Test Samples. The L2 samples (low-level samples run in between two low-level samples) would be categorized as Other, and L3 samples (low-level samples run last in each sequence) as Low Control Samples.
If you are following some other protocol, you can categorize your samples based on that. For example, if you are supposed to use only the last high-level samples of each sequence in calculating Hmean, then you should categorize those samples as High Level Samples and other high-level samples as Other. And if you are supposed to use both L2 and L3 samples in calculating L3mean, then you should categorize them all as Low Control Samples.
If your sequence varies but you still want to use the Cofrac equation in calculating the carryover, you can plan the protocol by giving the total number of high-level samples and the total number of low-level samples and setting the number of test runs as 1.
In addition to the Cofrac carryover, Validation Manager calculates absolute carryover as L1mean – L3mean, using the categorized samples as described above.
Mode 2: The custom protocol
Some carryover protocols use a varying sequence of high and low samples. The custom mode is specifically designed for these protocols, but you can just as well use it for protocols where a short sequence is repeated multiple times. When planning your protocol, simply give the total number of high-level samples and the total number of low-level samples. After importing your results, categorize your samples.
-
High Level Samples: samples in this category are not used in calculations, but you can see their results on the report to visually assess if their results are on the expected level, and whether the protocol you measured matches the sequence you were supposed to measure.
-
Low Test Samples: samples in this category are used in calculating LowTestmean i.e. the mean of the samples run directly after high-level samples.
-
Low Control Samples: samples in this category are used in calculating LowControlmean i.e. the mean of the samples to which the low test samples are compared.
-
Other: samples in this category are not used in calculations, but you can see their results on the report to visually assess if their results are on the expected level, and whether the protocol you measured matches the sequence you were supposed to measure.
Absolute carryover is calculated as LowTestmean – LowControlmean, and the relative carryover as
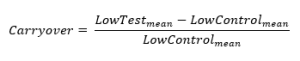 Try out the new study and tell us what you think about it. If any questions arise, we’d love to hear them.
Try out the new study and tell us what you think about it. If any questions arise, we’d love to hear them.
Accomplish more with less effort
See how Finbiosoft software services can transform the way your laboratory works.